Life Sciences
3D Spatial Colorectal Cancer Maps Combine Molecular and Histological Features
The Harvard Medical School maps are part of the team’s broader efforts to create atlases for different cancer types, which will be freely available to…
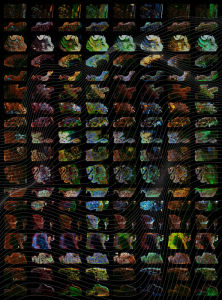
Researchers at Harvard Medical School have combined histology with cutting-edge single-cell imaging technologies to create large-scale 2D and 3D spatial maps of colorectal cancer. The maps layer extensive molecular information on top of histological features to provide new information about the structure of the cancer, as well as how it forms, progresses, and interacts with the immune system.

The maps are part of the team’s broader efforts to create atlases for different cancer types, which will be freely available to the scientific community as part of the National Cancer Institute’s Human Tumor Atlas Network. Previously, the researchers used a similar approach to create in-depth maps of early-stage melanoma, and maps for other cancers are in development. Ultimately, the team hopes that these cancer atlases will propel research and improve diagnosis and treatment.
“Our approach provides a molecular window into 150 years of diagnostic pathology — and reveals that many of the elements and structures traditionally thought to be isolated are actually interconnected in unexpected ways,” said Peter Sorger, PhD, the Otto Krayer Professor of Systems Pharmacology in the Blavatnik Institute at HMS. “An analogy is that before we were just looking at the tail or the foot of the elephant, but now, for the first time, we can start to see the whole elephant at once.” Sorger is co-senior author of the team’s study to map colorectal cancer, which is detailed in Cell, in a paper titled “Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer.”
Colorectal cancer is relatively slow growing and can often be treated surgically if caught early. However, treatment becomes more difficult if the disease progresses undetected, making it the fourth leading cause of cancer-related deaths in the U.S.
Despite the availability of colorectal cancer screening programs, treatment decisions for individual patients are still largely guided by traditional histology—colorectal cancer is assessed by examining slides of tumor samples under a microscope.
Histology has long been the cornerstone of cancer diagnosis and treatment. Pathologists examine a tumor sample stained with hematoxylin and eosin (H&E) under a microscope and pick out key features to determine the grade and stage of the cancer. This information is used by oncologists to develop a treatment plan, which usually involves some combination of surgery, drugs, and radiation. H&E-based histology is relatively simple, cheap, fast, and can reveal a lot about a tumor.
“Our existing maps of colorectal cancer originate in pathology—over the course of 150 years, we’ve figured out the most important H&E features for diagnosing a patient,” said co-senior author Sandro Santagata, PhD, HMS associate professor of systems biology and associate professor of pathology at Brigham and Women’s Hospital.

However, traditional histology has its limits. It doesn’t capture a cancer’s molecular makeup or physical structure, which makes it difficult to fully take advantage of the information cancer researchers have gained over the past 50 years. “Histology is extremely powerful, but we often don’t know what it means in modern molecular terms,” Sorger said. As the authors further pointed out, “ … classical methods provide insufficient information for mechanistic studies and precision medicine … Understanding intra-tumor heterogeneity (ITH) is essential for improving our knowledge of tumor biology and for optimizing diagnosis and therapy.”
Spatial tumor atlases aim to build on this foundation and modern genetic studies, by collecting detailed molecular and morphological information on cells captured in 3D environments. “Atlas construction is made possible by new highly multiplexed tissue imaging methods …” the scientists wrote.
For their newly reported work the team combined histology with single-cell molecular imaging data acquired through a multiplexed imaging technique called cyclic immunofluorescence, or CyCIF. They used this information to create detailed 2D maps of large regions of colorectal cancer. First author Jia-Ren Lin, PhD, platform director in the Laboratory of Systems Pharmacology at HMS, led the effort to stitch these maps together to form a large-scale 3D reconstruction of a tumor. “We use highly multiplexed tissue imaging, 3D reconstruction, spatial statistics, and machine learning to identify cell types and states underlying morphological features of known diagnostic and prognostic significance in colorectal cancer,” the investigators explained. “We show that accurate assessment of disease-relevant tumor structures requires the statistical power of whole-slide imaging (WSI), not the small specimens found in tissue microarrays (TMAs).”

“Our maps include information on almost 100 million cells from large pieces of tumors, and provide a rather unprecedented look at colorectal cancer,” Santagata said. The maps will allow researchers to start asking key questions about differences between normal and tumor tissues and variation within a tumor, he added, and reveal “exciting architectural features that had never been observed before, as well as molecular changes associated with these features.”
The resulting maps showed that a single tumor can have more and less invasive sections, and more or less malignant-looking regions—resulting in histological and molecular gradients where one part of a tumor transitions into the next. “Within each tumor, there is a wide range of properties of colorectal cancer—we see many different regions and neighborhoods that have distinct characteristics, as well as the transitions between them,” Santagata stated. The authors further noted, “Using 3D reconstruction of serial sections and supervised machine learning, we show that archetypical CRC histologic features are often graded and substantially larger than they appear in 2D.”
From here, Santagata added, scientists can start to explore what drives the differences within individual tumors. For example, the maps showed that immune environments varied dramatically within a single tumor. “They were as different across a single tumor as among tumors—which is important because tumor-immune interactions are what you are trying to target with immunotherapy,” Sorger commented.
“We find that the immune environment can vary substantially within a single tumor and recurrently with margin morphology across specimens,” the authors further stated. “At the tumor invasive margin, where tumor, normal, and immune cells compete, T cell suppression involves multiple cell types and 3D imaging shows that seemingly localized 2D features such as tertiary lymphoid structures are commonly interconnected and have graded molecular properties … Budding regions are the most T cell-rich, but also the most immunosuppressive (with abundant Tregs and PDL1-expressing cells). “Whereas tumor buds have few proliferating cells, tumor cells in deep invasive margins are highly proliferative and have fewer immediately adjacent immune cells.”
Similar to their finding in melanoma, the researchers observed that the T cells tasked with fighting off the cancer were not directly suppressed by tumor cells, but rather by other immune cells in the environment around the tumor. “This gives us a whole new appreciation for how diverse and plastic the tumor environments are—they are rich communities, and we are now better equipped to figure out how they develop,” Santagata said.
The maps also provided new insights into the architecture of the tumors. Scientists had previously identified what they thought were 2D pools of a mucus-like substance called mucin with clusters of cancer cells floating inside. However, in the new study, the 3D reconstruction revealed that these mucin pools are, in fact, a series of caverns interconnected by channels, with finger-like projections of cancer cells. “In 2D views, mucin surrounding bud-like structures is found in pools that appear isolated from each other,” the scientists stated. “In 3D, however, these mucin pools were frequently continuous with each other.” The intertumoral mucin pools, they noted, thus comprise “ … 3D networks that can connect to the intestinal lumen and its microbiome.”
“It’s a wild, new look at these tumor structures that we never really appreciated before,” Santagata said. “Because we can see them in 3D, we have a crisp, clean view of the structures, and we can now study why they are there, how they form, and how they shape tumor evolution.” The authors further noted “Thus, while cancer genetics emphasizes the importance of discrete changes in tumor state, whole-specimen imaging reveals large-scale morphological and molecular gradients analogous to those in developing tissues.”
Ultimately, the goal of these colorectal cancer and other cancer maps in development is to advance research and improve diagnosis and treatment. Precision medicine, which involves tailoring therapy to an individual patient’s cancer, is becoming an increasingly important part of treatment, Sorger noted, yet it can go only so far with pathology and genetics alone.
“The big translational story here is building the knowledge to make precision medicine practical for most patients,” he said. “We are currently working with Brigham and Women’s and the Dana-Farber Cancer Institute to determine how our methods can be used in a clinical setting.”
“This is allowing us to extract a whole additional layer of molecular and structural features that we think will provide diagnostic and prognostic information and improve our ability to target these cancers,” Santagata added.
The researchers want to further refine their ability to create 3D reconstructions of tumors and continue integrating new imaging technologies into their maps. They also want to build a bigger cohort of colorectal cancer samples for mapping and explore the basic biology of the disease that their maps have highlighted. One limitation of the reported study, they acknowledged, is that only one CRC has yet been constructed in 3D, “largely because the process remains manual and slow, and many of the features we describe in 3D—TB fibrils, TLS networks, and invasive margins—would benefit from deeper molecular profiling to better identify cell types and states.” In addition, they noted, “There are many spatial relationships among the 2 x 108 cells in our dataset that we have not yet explored.”
For Sorger, the project represents an unusual collaboration between pathologists, engineers, and computational scientists: As the imaging data rolled in, the computational scientists used machine learning to identify interesting findings that they presented to the pathologists, and the pathologists flagged key features to be parsed with machine learning.
“This was an extraordinarily close conversation between the computational group and the pathology group, going back and forth between the rich history of medicine known to pathologists and modern machine learning methods.” Sorger said. “I think it’s an exciting glimpse of how these computation methods can be used in medicine in the future, wherein you tightly couple biologists and physicians with computation, rather than seeing them as replacements for each other.”
The team chose melanoma and colorectal cancer as a starting point because they are common cancers with unmet medical needs that consist of large, solid tumors and require important treatment decisions. Next, the researchers plan to tackle breast cancer and brain cancer. They also want to train other scientists to use the imaging technologies to build their own cancer maps, which would pave the way for the creation of even more atlases.
“A new era in molecular pathology is beginning, and this is a deep look at a tumor that is showing us how remarkable the findings can be,” Santagata stated.
The post 3D Spatial Colorectal Cancer Maps Combine Molecular and Histological Features appeared first on GEN – Genetic Engineering and Biotechnology News.
immunotherapy
microbiome
medical
machine learning
medicine
hospital

Wittiest stocks:: Avalo Therapeutics Inc (NASDAQ:AVTX 0.00%), Nokia Corp ADR (NYSE:NOK 0.90%)
There are two main reasons why moving averages are useful in forex trading: moving averages help traders define trend recognize changes in trend. Now well…
Spellbinding stocks: LumiraDx Limited (NASDAQ:LMDX 4.62%), Transocean Ltd (NYSE:RIG -2.67%)
There are two main reasons why moving averages are useful in forex trading: moving averages help traders define trend recognize changes in trend. Now well…
Asian Fund for Cancer Research announces Degron Therapeutics as the 2023 BRACE Award Venture Competition Winner
The Asian Fund for Cancer Research (AFCR) is pleased to announce that Degron Therapeutics was selected as the winner of the 2023 BRACE Award Venture Competition….














